Publications
Dissertations
 |
Ehsaneddin Asgari
Life Language Processing: Deep Learning-based Language-agnostic Processing of Proteomics, Genomics/Metagenomics, and Human Languages.
UC Berkeley, 2019
|
Journal Papers |
||||
|
|
 |
Asgari, E., Poerner, N., McHardy, A., & Mofrad, M.
bioRxiv, 705426 (2019).
Under Review.
|
||
 |
 |
Khaledi A*, Weimann A*, Schniederjans M§, Asgari E§, Kuo T, Gabriel O, Kola A, Gastmeier P, Hogardt M, Jonas D, Mofrad MRK, Bremges A, McHardy AC, Hussler S.
EMBO Molecular Medicine 2020 e10264 . |
||
 |
Zhou, N., Jiang, Y., Bergquist, T. R., Lee, A. J., Kacsoh, B. Z., Crocker, A. W., … & Davis, L.
Genome Biology 20, 244 (2019) doi:10.1186/s13059-019-1835-8. |
|||
|
DiMotif ProtVecX |
|
 |
Asgari, E, McHardy A. C., and Mofrad M. R. K.
Scientific reports 9.1 (2019): 3577.
|
|
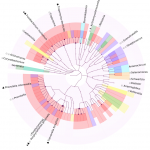
DiTaxa |
 |
Asgari, E, Münch P. C., Lesker T.R., McHardy A. C., and Mofrad M. R. K.
Bioinformatics (2018) bty954, https://doi.org/10.1093/bioinformatics/bty954.
|
||
 |
|
 |
Asgari E, Garakani K, McHardy AC and Mofrad M. R. K,
Bioinformatics, Volume 34, Issue 13, 1 July 2018, Pages i32–i42, https://doi.org/10.1093/bioinformatics/bty296
ISMB 2018 Special Issue: microbial informatics track — acceptance rate fo 0.13
|
|

BioVec ProtVec GeneVec |
Asgari, E, and RK Mofrad, MRK.
Continuous distributed representation of biological sequences for deep proteomics and genomics. PloS ONE 10.11 (2015): e0141287. |
|||
Peer-reviewed Conference/Workshop Papers |
||||
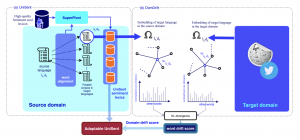 |
E. Asgari, F. Braune, B. Roth, C. Ringlstetter, M. R.K. Mofrad,
UniSent: Universal Adaptable Sentiment Lexica for 1000+ Languages. In Proceedings of Language Resources and Evaluation Conference (LREC) 2020 , Marseille, France, May 2020. |
|||
| Highlighted by MIT Tech. Review Magazine
|
E. Asgari, H. Schütze ,
Past, Present, Future: A Computational Investigation of the Typology of Tense in 1000 Languages. In Proceedings of the Empirical Methods on Natural Language Processing (EMNLP), Copenhagen, Denmark, September 2017. |
|||
| BEST PAPER AWARD
Sponsored by: |
E. Asgari, M.R.K. Mofrad,
In Proceedings of the NAACL-HLT Workshop on Multilingual and Cross-lingual Methods in NLP, San Diego, CA, 2016. |
|||
| Selected among best submissions
|
E. Asgari, M.R.K. Mofrad,
Word Vectors of Biological Sequences and Their Applications in Bioinformatics, International Machine Learning Conference (ICML) Workshop on Computational Biology, New York City, NY, 2016. |
|||
| E. Asgari, S. Nasiriany, M.R.K. Mofrad,
Text Analysis and Automatic Triage of Posts in a Mental Health Forum, In Proceedings of the NAACL-HLT Workshop on Computational Linguistics and Clinical Psychology, San Diego, CA, 2016. |
||||
Book chapters |
||||
| E. Asgari and M. R. K Mofrad Deep Genomics and Proteomics: Language Model-Based Embedding of Biological Sequences and Their Applications in Bioinformatics Leveraging Biomedical and Healthcare Data (pp. 167-181) 2019. Academic Press. |
||||
| H. Adel and E. Asgari and H. Schütze Overview of Character-Based Models for Natural Language Processing Lecture Notes in Computer Science book series (LNCS, volume 10761) 2018, 3–16. |
||||